

What is the reference splicing pattern.
What nomenclature is used for introns?
Does a particular database name have any meaning?
What is the id used in the Exon and intron FASTA files?
Q: What is the reference splicing pattern.
The reference splicing pattern is used to apply a numbering system on the exons based on the numbering of exons in the first known splicing pattern detected for a particular gene. The reference splicing pattern is chosen as the most recently updated RefSeq with the highest version number; normally the higher the version number the longer a RefSeq accession has been around. Exceptionally a reference splicing pattern will be imposed to avoid awkward numbering of exons.
Q: Why are some exons marked as being constitutive (‘C’ in the Exon table) and yet do not appear on any RefSeq isoform.
Exons marked as constitutive are those which are not detected as being spliced out by a splicing reaction; first or terminal exons will only be marked as constitutive if the gene contains a single first or single terminal exon respectively.
Exons marked as constitutive that do not appear on any RefSeq isoform will occur only in the case of exons which are upstream or downstream of the reference splicing pattern: in the case of more than one novel exon upstream or downstream, i.e. indicating possible alternative promotors or alternative poly-adenylation; or in the rare case of splicing detected to another upstream or downstream gene, in which case they will be constitutive exons belonging to that other gene. These exons can be distinguished from the exons present in the reference splicing pattern by their reference numbering – the reference number for exons upstream or downstream of the reference splicing pattern is preceded by the capital letter U or W respectively.
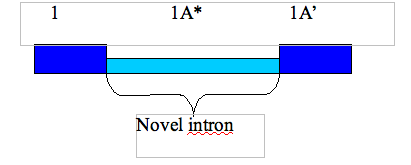
Q: What nomenclature is used for introns?
An intron is refered to using the numbers of the flanking exons separated by an underscore, e.g. the intron lying between exon 1 and 2 is refered to as intron 1_2. In the case of a novel intron, i.e. one that is not spliced out in the reference sequence, the reference nomenclature used is as shown below:
Q: Does a particular database name have any meaning?
Yes, the database are named using a code which can quickly give us the month and year of the update, the organism, and the version of the program which was used to run the data.
e.g. d07hs60 (December 2007, Homo sapiens, ExonMine version 60)
for the organism a two letter code is used:
Human - Homo sapiens (hs)
Chimp - Pan troglodytes (pt)
Rhesus - Macaca mulatta (rm)
Cow - Bos taurus (bt)
Dog - Canis lupus familiaris (cf)
Rat - Ratus norvegicus (rn)
Mouse - Mus musculus (mm)
Zebrafish - Danio rerio (dr)
Chicken - Gallus gallus (gg)
Frog - Xenopus tropicalis (xt)
Fly - Drosophila melanogaster (dm)
Nematode worm - Caenorhabditis elegans (ce)
Sea squirt - Ciona intestinalis (ci)
For the months the following code is used: Jan(j), Feb(f), Mar(r), Apr(a), May(m), June(u), July(y), Aug(g), Sept(s), Oct(t), Nov(n), Dec(d).