These are some of the projects being developed in the Lab, with additional [keywords] highlighting our main interests:
RNA (de)regulation in disease contexts
Cancer
The role of tumour-infiltrating myeloid cells in breast
cancer at the single-cell level [immuno-senescence, scRNA-seq analysis]
Lab members involved: Marta Bica, Nuno Barbosa-Morais (Co-PI)
Collaborators: Karine
Serre (PI)
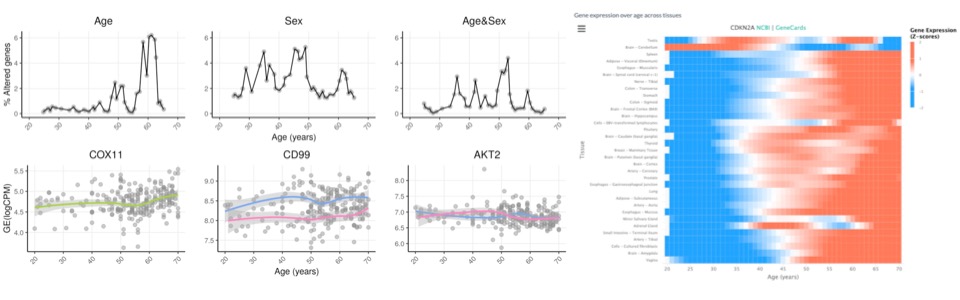
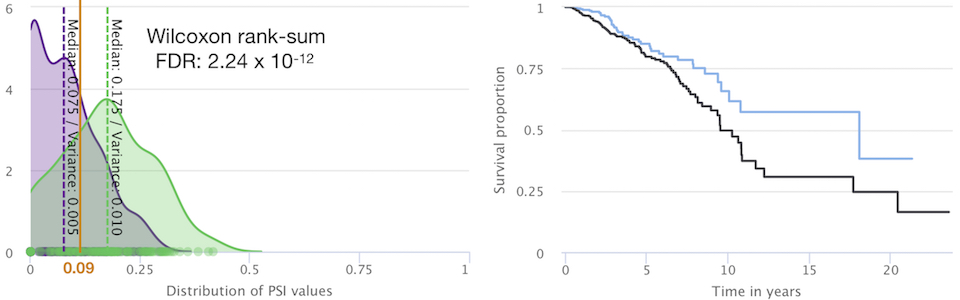
CDKN2A in senescence and cancer [alternative splicing, senescence]
Lab members involved: Rita Silva, Maria Guerra, José
Ferrão (alumnus), Mariana Ascensão-Ferreira, Nuno Barbosa-Morais (PI)
Collaborators: Maria
Vivo
Uncovering the stressome: a computational approach to define a stress granule signature and its implication in cancer
Lab members involved: Alexandre Kaizeler, Nuno Barbosa-Morais (PI)
Alternative transcription of CELF2 in colorectal cancer
progression and therapeutics [alternative splicing]
Lab members involved: Francisca Xara-Brasil, Lina Gallego (alumna) (Co-PI), Marta Bica, Nuno Barbosa-Morais (PI)
Collaborators: Marta
Martins (Co-PI), Luís
Costa, Ana Rita Grosso
Alternative splicing during oncogene-induced senescence [alternative splicing, senescence]
Lab members involved: Mariana Ascensão-Ferreira, Nuno Barbosa-Morais (Co-PI)
Collaborators: Jesús
Gil (PI)
Moreno-Marin N, et al. bioRxiv, 2023.03.13.532472.
Conde J, et al. ACS Central Science, 2021 April 14;7(5):868–881. [PUBMED]
Gomes I, et al. Oncotarget, 2020 May 12;11(19):1714-1728. [PUBMED]
Munkley J, et al. eLife, 2019 Sep 3;8:e47678. [PUBMED]
Rodrigues T, et al.Chem Commun (Camb), 2019 May 30;55(45):6369-6372. [PUBMED]
Baker C, et al. Bioorg Med Chem, 2019 Jun 15;27(12):2531-2536. [PUBMED]
de Almeida BP, et al. PLoS Computational Biology, 2019 Mar 11;15(3):e1006832. [PUBMED]
Braun S, et al. Nat Commun, 2018 Aug 17;9(1):3315. [PUBMED]
Georgilis A, et al. Cancer Cell, 2018 Jul 9;34(1):85-102.e9. [PUBMED]
Marteil G, et al. Nat Commun, 2018 Mar 28;9(1):1258. [PUBMED]
Neuro-biology
Investigating circadian disruption in a submarine crew: the health impact of alternating shift work in low-light and confined environments
Lab members involved: Daniel Marques, Nuno
Barbosa-Morais (PI)
Collaborators: Cátia
Reis (Co-PI)
Sousa NS, et al. bioRxiv, 2023.11.07.565995, Cell Reports (in press).
Martins I, et al. Int J Mol Sci, 2023;24(7):6433. [PUBMED]
Goulielmaki E, et al. Nature Communications, 2021 May 26;12(1):3153. [PUBMED]
Bordone MC & Barbosa-Morais NL. Frontiers in Neuroscience, 2020 Dec 9;14:607215. [PUBMED]
Rathore OS, et al. RNA, 2020 Dec;26(12):1935-1956. [PUBMED]
Godinho-Silva C, et al. Nature, 2019 Oct;574(7777):254-258. [PUBMED]
Leznicki P, et al. J Cell Sci. 2018 May 16;131(10):jcs.212753. [PUBMED]
Cardoso V, et al. Nature, 2017 Sep 14;549(7671):277–281. [PUBMED]
Gallego-Paez LM, et al. Human Genetics, 2017 Sep;136(9):1015-1042. [PUBMED]
Braunschweig U, et al. Genome Res, 2014 Nov;24(11):1774-86. [PUBMED]
Han H, et al. Nature, 2013 Jun 13;498(7453):241-5. [PUBMED]
Ward MC, et al. Mol Cell, 2013 Jan 24;49(2):262-72. [PUBMED]
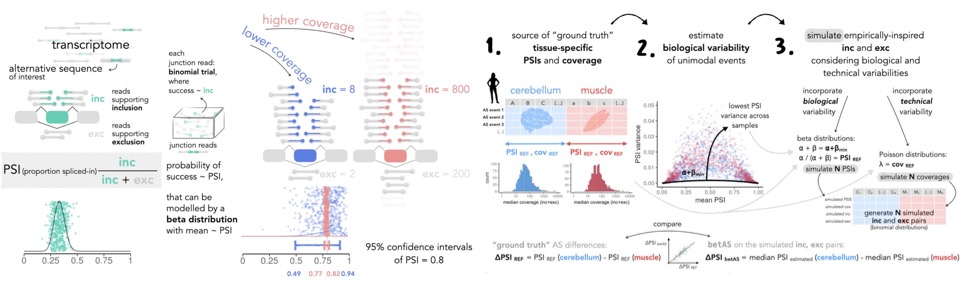
Bioinformatics tools for the analysis of RNA-seq data
cTRAP:
identification of candidate causal perturbations from differential
gene expression data [pharmaco-transcriptomics]
Lab members involved: Bernardo
Almeida (alumnus), Nuno Saraiva-Agostinho (alumnus) (co-PI), Juan
Carlos Gómez Verjan (alumnus), Nuno Barbosa-Morais (PI)
scStudio: Shiny-based interactive graphical-interface for
the exploration of pre-processed single-cell transcriptomics data
[immuno-senescence, scRNA-seq analysis]
Lab members involved: Marta Bica, Nuno Barbosa-Morais (PI)
Collaborators: Karine
Serre, Michael Gotthardt
Schneider AL*, Martins-Silva R*, Kaizeler A*, Saraiva-Agostinho N, Barbosa-Morais NL. eLife, 2024;12:RP88623. [PUBMED]
Ascensao-Ferreira M*, Martins-Silva R*, Saraiva-Agostinho N, Barbosa-Morais NL. RNA, 2024;30(4):337-353. [PUBMED]
Saraiva-Agostinho N & Barbosa-Morais NL. Methods in Molecular Biology, 2020;2117:179-205. [PUBMED]
Saraiva-Agostinho N & Barbosa-Morais NL. Nucleic Acids Research, 2019 Jan 25;47(2):e7. [PUBMED]
Funding