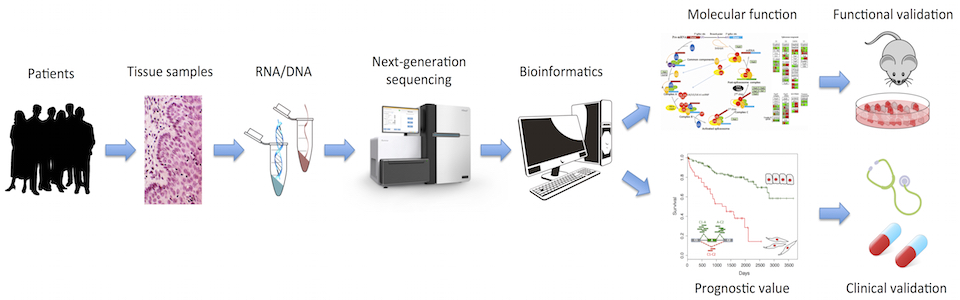
The Bioinformatics Matters Community and GenomePT

Bioinformatics Matters is a Research Community born due to the need of gathering all iMMers with an interest in Bioinformatics. It aims to consolidate the fundamental FAIR principles, contributing towards the unity of wet-lab researchers, and dry-lab researchers, by making bioinformatics knowledge available to the entire scientific community. It is focused on three main pillars: sharing, training, and networking.
GenomePT is a consortium of Portuguese research infrastructures, including iMM, that aims to provide sequencing and bioinformatics services for genome projects. GenomePT is integrated in the Portuguese Roadmap of Research Infrastructures.
The GenomePT iMM Node supports the Bioinformatics Matters community and its activities, by directly funding iMM researchers to make their work in the bioinformatics field accessible to the national research community.
POCI-01-0145-FEDER-022184, supported by COMPETE 2020 – Operational Programme for Competitiveness and Internationalisation (POCI), Lisboa Portugal Regional Operational Programme (Lisboa2020), Algarve Portugal Regional Operational Programme (CRESC Algarve2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).