User Tools
![]() STaQTool - Spot Tracking and Quantification Tool
STaQTool - Spot Tracking and Quantification Tool
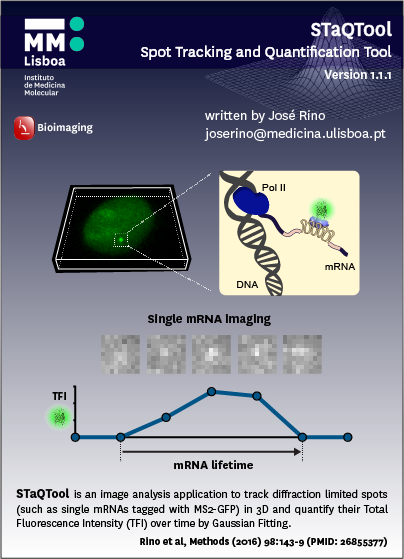
STaQTool was developed for automated tracking of a transcription site in the cell nucleus, measuring its fluorescence intensity by Gaussian fitting and analyzing patterns of changes in fluorescence intensity over time. STaQTool runs in any PC with MATLAB Compiler Runtime. The software is a set of open-source MATLAB functions integrated into a single GUI which integrates all functions in a self-explanatory interface and allows for data visualization and easy adjustment of tracking and fitting parameters, as well as automated calibration procedures and analysis of either single transcription sites or multiple mRNA particles moving throughout the nucleus or the cytoplasm. Results such as coordinates of the transcription site and total fluorescence intensity (TFI) are saved to Excel files automatically and allow the user to pause and resume analysis at any time, as well as set up batch analysis of multiple files for unattended automated processing.
 |
→ (installs MATLAB Compiler Runtime 9.0.1 if not present) |
 setup file for Windows x64
setup file for Windows x64